
“FISH” (Fluoresence in-situ Hybridization) is a powerful tool for investigating the relative abundance and physical characteristics of different populations of microorganisms.
FISH combines fluorescence microscopy with oligonucleotide probes--precisely designed DNA sequences conjugated with fluorescent molecules that are used to “tag,” visualize and quantify specific microbes of interest.
Geovation’s multi-color FISH (“mFISH”) lab uses multiple probes with different color fluorochromes to simultaneously provide quantitative information on different sub-populations in the same sample.
Geovation offers several mFISH assays to provide relatively rapid and cost-effective enumeration of important groups of microorganisms relative to other methods. mFISH also enables the simultaneous visualization of the spatialarrangements of microbes in mixed consortia to investigate the potential for mutualistic or syntrophic interactions among different organisms.
FISH provides data useful for applications ranging from biological monitoring of monitored natural attenuation (”MNA”) programs, bioremediation monitoring and process optimization, bacterial source tracking (”BST”), medical and public-health microbiology, and problem solving and process monitoring for waste-water treatment.
|
Capabilities & Utility of Geovation's mFISH Lab Services
Quantify total microbial cells, specific microbes and groups of microbes at different taxonomic levels: Spatial mapping of population abundance Evaluate shifts in target populations over time Detection limits: approx. 104 cells / mL (2 mL sample aliquot) Investigative problem-solving in microbial ecology: Bioremediation and MNA programs |
||||||||||||
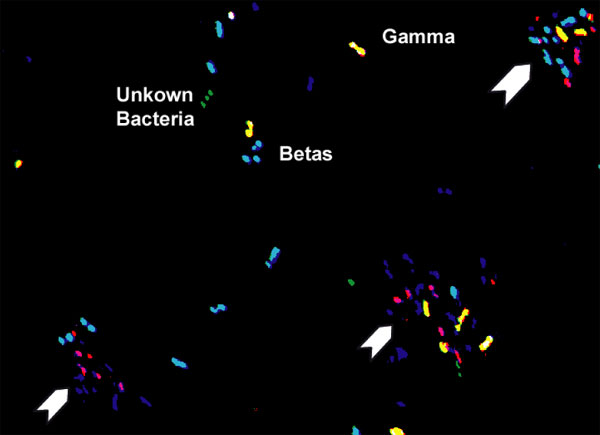
False-color RGB composite bitmap image of a "MP1" mFISH assay showing denitrifying bacteria from a gasoline-contaminated aquifer. Aqua cells are beta-proteobacteria (dual hybridizations with 'blue' BET42a and 'green' EUB338 probes), yellow cells are gamma- proteobacteria (dual hybridizations with 'red' GAM42a and 'green' EUB338 probes) and green cells are undifferentiated bacteria (single hybridizations with 'green' EUB338 probe). Red, purple, and blue objects are noise from cells within biofilms (arrows). |
||||||||||||
| Geovation's FISH Lab Fast-Facts - What ís it all about?
Procedures Sterile sample vials with fixative / preservative are shipped to the client under standard chain-of-custody procedures Water and ground-water samples are collected and “fixed” in the field, kept cool and returned via overnight courier to Geovation (soil samples or samples containing solids require special handling and preparation procedures - additional charges may apply) In the hybridization lab, samples are filtered, hybridized with fluorochrome-labeled oligonucleotide probes (multiple probes & colors for mFISH assays), washed, DAPI stained and mounted onto slides A computer-controlled fluorescence microscope is used to visualize and CCD image the samples for subsequent computer-assisted, semi-automated digital image processing, cell counting & classification |
||||||||||||

Evidence of Syntrophism? |
||||||||||||
| FISH Data / Reporting
Reports are provided in Excel or pdf format with data for each target group reported in units of cells/mL and % of DAPI counts. Each FISH assay provides a minimum of 2 to 4 “data points” per sample -- total cell counts (DAPI staining) along with the cell counts and relative percentages of DAPI counts for each target population. Several “mFISH” assays are available that provide simultaneous data for 2-4 target populations plus total cell counts (DAPI). |
||||||||||||
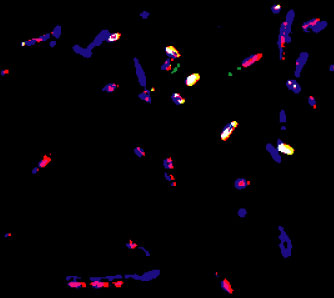
Abundant Archaea? Tot. Bacteria (Red-Violet) and beta-proteobacteria (Yellow-White) cells are differentiated from other DAPI-stained cells (Blue) that did not hybridize with either the EUB338 or BET42a probes. False-color composite image from a "MP1" mFISH assay performed on a ground-water sample from a TCE-contaminated aquifer. Bacteria represent about 70% of total DAPI-stained cells, beta-proteobacteria about 30%. Many of the 30% of DAPI-stained cells that did not hybridize with EUB338 may be archaea. Little is currently known about psychrotolerant archaea in the environment and their role in biodegradation and bioremediation. |
||||||||||||
| Examples of FISH Assays / Parameters
High-Level Taxonomic Groups Total bacteria; alpha-, beta-, gamma-, and delta-proteobacteria; CFB (cytophaga-flexibacter-bacteroides); high and low GC gram+ bacteria Total Archaea; Crenarcheota; Euarcheota; Total yeast / fungi Targeted Groups - Family / Genus / Species Levels Important subgroups of proteobacteria Pseudomonas spp., Bacteroides spp., Bfidobacterium spp. Dehalococcoides ethenogenes (DHE) Multi-parameter mFISH Assays MP1 Tot. DAPI; Tot. bacteria; Tot. beta- and gamma-proteobacteria MP3 Tot. DAPI; Tot. bacteria; total yeast MP5 Tot. DAPI; Tot. CFB; DHE; delta-proteobacteria MP7 Tot. DAPI; Tot. archaea; Tot. crenarcheota; Tot. euarcheota MP8 "BST" applications - Tot. DAPI; human-related CFB; ruminant-related Fibrobacter spp.; Campylobacter spp. (geese) Many probes and several "MP" multi-color, mFISH probe combinations available. Custom assays available on request. |
||||||||||||
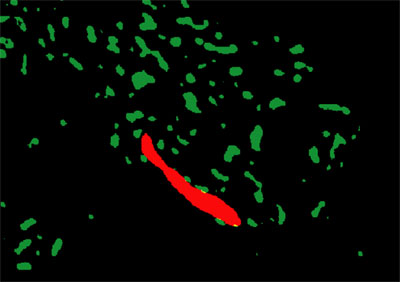
Dimorphic yeast (Red) among bacterial cells (Green) showing through biofilm EPS (Black - removed); MP3 assay, gasoline plume |
||||||||||||
|
© 2012 Geovation Engineering, P.C.. All Rights Reserved.
2016 Route 284 • P.O. Box 513 • Slate Hill, New York 10973 • (845) 697-5100 |
||||||||||||